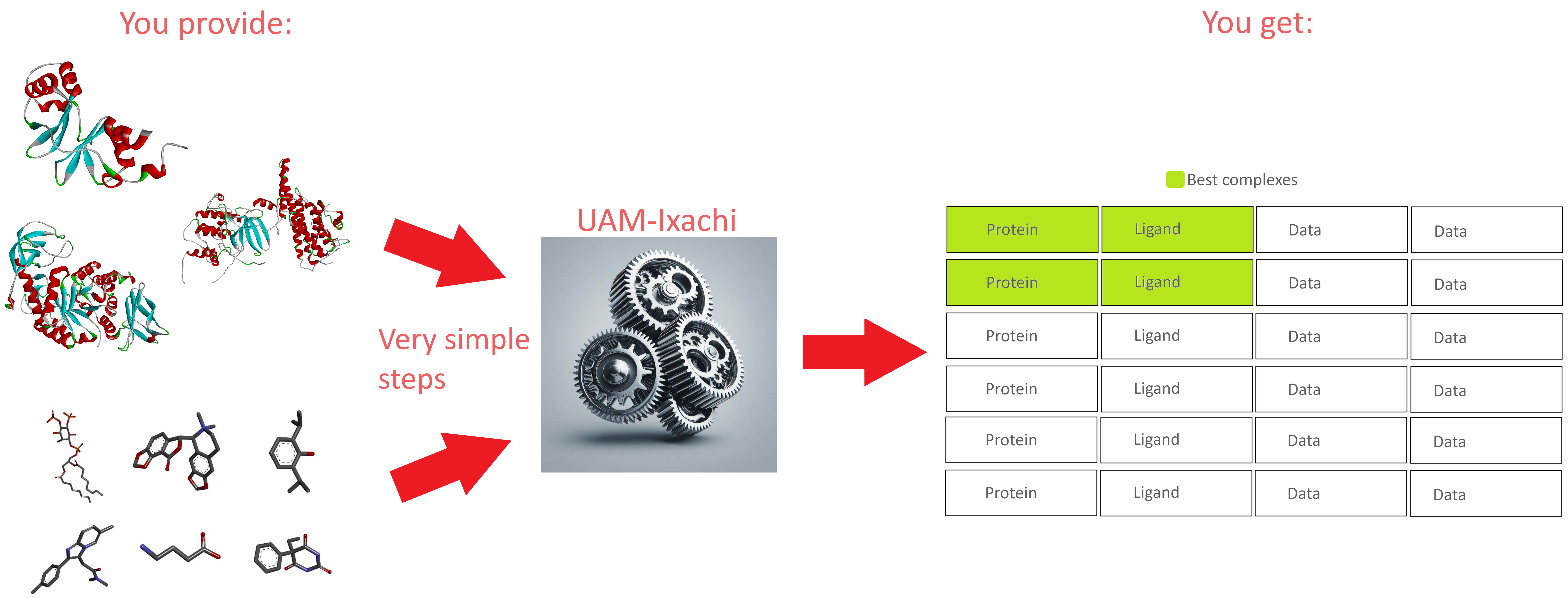
UAM-Ixachi: Desktop Tool for Massive Automated Molecular Docking
DOI:
https://doi.org/10.29356/jmcs.v69i1.2299Keywords:
Molecular docking, virtual screening, CADD, in silico simulation, drug designAbstract
The molecular docking has become a powerful computational tool for new drug research and design, playing a key role in predicting interactions between drug-related ligands and their potential target proteins. However, molecular docking and virtual screening simulation software currently available require researchers to make numerous configurations and navigate unintuitive menus, necessitating significant process optimization. The present work used existing tools for molecular docking, designing a set of coherent computational programs among themselves, with the aim of expediting work with many ligands and target proteins, and simplifying the simulations performed simultaneously, making these techniques accessible to researchers with limited computational skills. The aim was to design an open-source tool, free and simple to use for the academic community, through the URL: https://1drv.ms/f/s!AiwrqGMGvesstXgOcz3Hn1Q2mfI9?e=903be7, offering a robust format for the presentation of results, conceptualized as a massive report of rows and columns that facilitates the management and interpretation of a large amounts of data.
Resumen. La simulación de acoplamiento molecular se ha convertido en una poderosa herramienta computacional para el descubrimiento y diseño de fármacos, desempeñando un papel fundamental en la predicción de las interacciones de unión entre ligandos de interés farmacológico y sus dianas potenciales. Sin embargo, los programas de simulación de acoplamiento molecular y cribado virtual disponibles en la actualidad requieren que los investigadores realicen numerosas configuraciones y naveguen por menús poco intuitivos, lo que hace necesario eficientizar y acelerar significativamente este proceso. Este trabajo utilizó las herramientas existentes para simulación de acoplamiento molecular, para diseñar un conjunto de programas computacionales coherentes entre sí, buscando agilizar el trabajo con una gran cantidad de ligandos y proteínas, y simplificar las simulaciones realizadas simultáneamente, facilitando el acercamiento de estas técnicas a investigadores poco instruidos en informática. El objetivo fue diseñar una herramienta de código abierto, gratuito y simple de usar para la comunidad académica, a través de la URL https://1drv.ms/f/s!AiwrqGMGvesstXgOcz3Hn1Q2mfI9?e=903be7, ofreciendo un formato robusto de presentación de resultados, conceptualizado como un reporte masivo de filas y columnas que facilita el manejo y la interpretación de la gran cantidad de datos obtenidos.
Downloads
References
Prieto-Martínez, F. D.; Arciniega, M.; Medina-Franco, J. L. Tip Rev. Esp. Ciencias Químico-Biol. 2018, 21. DOI: https://doi.org/10.22201/fesz.23958723e.2018.0.143. DOI: https://doi.org/10.22201/fesz.23958723e.2018.0.143
Shoichet, B. K.; McGovern, S. L.; Wei, B.; Irwin, J. J. Curr. Op. Chem. Biol. 2002, 6, 439-446. DOI: https://doi.org/10.1016/s1367-5931(02)00339-3. DOI: https://doi.org/10.1016/S1367-5931(02)00339-3
Bello, M. Rev. Digital Universitaria. 2021, 22, 5. DOI: https://doi.org/10.22201/cuaieed.16076079e. DOI: https://doi.org/10.22201/cuaieed.16076079e.2021.22.6.5
Kitchen, D. B.; Decornez, H.; Furr, J. R.; Bajorath, J. Nat. Rev. Drug Discov./Nat. Rev. Drug Disc. 2004, 3, 935-949. DOI: https://doi.org/10.1038/nrd1549. DOI: https://doi.org/10.1038/nrd1549
Stanzione, F.; Giangreco, I.; Cole, J. C., in: Progress in Medicinal Chemistry. 2021, 60, 273-343. DOI: https://doi.org/10.1016/bs.pmch.2021.01.004. DOI: https://doi.org/10.1016/bs.pmch.2021.01.004
Morris, G. M.; Lim-Wilby, M., in: Methods in Molecular Biology. 2008, 443, 365-382. DOI: https://doi.org/10.1007/978-1-59745-177-2_19. DOI: https://doi.org/10.1007/978-1-59745-177-2_19
Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. J. Mol. Biol. 1996, 261, 470-489. DOI: https://doi.org/10.1006/jmbi.1996.0477. DOI: https://doi.org/10.1006/jmbi.1996.0477
Brooijmans, N.; Kuntz, I. D. Ann. Rev. Bioph. Biomol. Struct. 2003. 32, 335-373. DOI: https://doi.org/10.1146/annurev.biophys.32.110601.142532. DOI: https://doi.org/10.1146/annurev.biophys.32.110601.142532
Morris, G. M.; Goodsell, D. S.; Halliday, R. S.; Huey, R.; Hart, W. E.; Belew, R. K.; Olson, A. J. J. Comp. Chem. 1998, 19, 1639-1662. DOI: https://doi.org/10.1002/(sici)1096-987x(19981115)19:14. DOI: https://doi.org/10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
Agu, P. C.; Afiukwa, C. A.; Orji, O. U.; Ezeh, E. M.; Ofoke, I. H.; Ogbu, C. O.; Ugwuja, E. I.; Aja, P. M. Sci. Rep. 2023, 13, 13398. DOI: https://doi.org/10.1038/s41598-023-40160-2. DOI: https://doi.org/10.1038/s41598-023-40160-2
Paucara, W. G. B.; Torrez, R. E. G. Rev. CON-CIENCIA. 2019, 7, 55-72.
Feinstein, W. P.; Brylinski, M. J. Cheminform. 2015, 7, 18. DOI: https://doi.org/10.1186/s13321-015-0067-5. DOI: https://doi.org/10.1186/s13321-015-0067-5
Singh, M.; Malhotra, L.; Haque, M. A.; Kumar, M.; Tikhomirov, A.; Litvinova, V.; Korolev, A. M.; Ethayathulla, A.; Das, U., Shchekotikhin, A. E.; Kaur, P. Biochimie. 2021, 182, 152-165. DOI: https://doi.org/10.1016/j.biochi.2020.12.024. DOI: https://doi.org/10.1016/j.biochi.2020.12.024
Guan, Y.; Jiang, S.; Ye, W.; Ren, X.; Wang, X.; Zhang, Y.; Yin, M.; Wang, K.; Tao, Y.; Yang, J.; Cao, D.; Cheng, Y. Cell Death Dis. 2020, 11, 948. DOI: https://doi.org/10.1038/s41419-020-03153-x. DOI: https://doi.org/10.1038/s41419-020-03153-x
Choowongkomon, K.; Sawatdichaikul, O.; Songtawee, N.; Limtrakul, J. Receptor- Mol./Molecules Online/Mol. Ann. 2010, 15, 4041-4054. DOI: https://doi.org/10.3390/molecules15064041. DOI: https://doi.org/10.3390/molecules15064041
Arba, M.; Ihsan, S.; Ramadhan, L. O. A. N.; Tjahjono, D. H. Comp. Biol. Chem. 2017, 67, 9-14. DOI: https://doi.org/10.1016/j.compbiolchem.2016.12.005. DOI: https://doi.org/10.1016/j.compbiolchem.2016.12.005
Asnawi, N. A.; Aman, L.; Nursamsiar, N.; Yuliantini, N. A.; Febrina, N. E. Rasayan J. Chem. 2022, 15, 1352-1361. DOI: https://doi.org/10.31788/rjc.2022.1526769. DOI: https://doi.org/10.31788/RJC.2022.1526769
Roy, S.; Ali, A.; Bhattacharya, S. J. Phys. Chem. B. 2021, 125, 5489-5501. DOI: https://doi.org/10.1021/acs.jpcb.0c10991. DOI: https://doi.org/10.1021/acs.jpcb.0c10991
Giacoman-Martínez, A.; Alarcón-Aguilar, F. J.; Zamilpa, A.; Huang, F.; Romero-Nava, R.; Román-Ramos, R.; Almanza-Pérez, J. C. Can. J. Physiol. Pharmacol. 2021, 99, 935-942. DOI: https://doi.org/10.1139/cjpp-2021-0027. DOI: https://doi.org/10.1139/cjpp-2021-0027
Dong, L.; Shen, S.; Chen, W.; Xu, D.; Yang, Q.; Lu, H.; Zhang, J. J. Chem. Inf. Model. 2019, 59, 4374-4382. DOI: https://doi.org/10.1021/acs.jcim.9b00479. DOI: https://doi.org/10.1021/acs.jcim.9b00479
Song, Y.; Lim, J.; Seo, Y. H. 2019. Eur. J. Med. Chem. 164, 263-272. DOI: https://doi.org/10.1016/j.ejmech.2018.12.056. DOI: https://doi.org/10.1016/j.ejmech.2018.12.056
Taherkhani, A.; Moradkhani, S.; Orangi, A.: Jalalvand, A.; Khamverdi, Z. Res. J. Pharmacognosy. 2021, 8, 37-51. DOI: https://doi.org/10.22127/rjp.2021.288366.1705.
Gentile, F.; Oprea, T. I.; Tropsha, A.; Cherkasov, A. Chem. Soc. Rev. 2003, 52, 872-878. DOI: https://doi.org/10.1039/d2cs00948j. DOI: https://doi.org/10.1039/D2CS00948J
Medina-Franco, J. L.; Martinez-Mayorga, K.; Gortari, E. F.; Kirchmair, J.; Bajorath, J. F1000Res. 2021, 10, 397. DOI: https://doi.org/10.12688/f1000research.52676.1. DOI: https://doi.org/10.12688/f1000research.52676.1
Saldívar-González, F. I.; Prieto-Martínez, F. D.; Medina-Franco, J. L. Educ. Quim. 2017, 28, 51-58. DOI: https://doi.org/10.1016/j.eq.2016.06.002. DOI: https://doi.org/10.1016/j.eq.2016.06.002
Ballón, W.; Grados, R. Rev. Cs. Farm. Y Bioq. 2019, 7, 55-72.
Morris, G. M.; Huey, R.; Lindstrom, W.; Sanner, M. F.; Belew, R. K.; Goodsell, D. S.; Olson, A. J. J. Comp. Chem. 2019, 30, 2785-2791. DOI: https://doi.org/10.1002/jcc.21256. DOI: https://doi.org/10.1002/jcc.21256
Forli, S. Raccoon AutoDock VS Preparation Tool. Center For Computational Structural Biology. 2009. https://autodock.scripps.edu/download/376/, accessed in July 2023.
Grosdidier, A.; Zoete, V.; Michielin, O. J. Comp. Chem. 2011, 32, 2149-2159. DOI: https://doi.org/10.1002/jcc.21797. DOI: https://doi.org/10.1002/jcc.21797
De Vries, S. J.; Van Dijk, M.; Bonvin, A. M. J. J. Nat. Prot. 2010, 5, 883-897. DOI: https://doi.org/10.1038/nprot.2010.32. DOI: https://doi.org/10.1038/nprot.2010.32
Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S. Nucleic Acids Res. 2017, 45, 365-373. DOI: https://doi.org/10.1093/nar/gkx407. DOI: https://doi.org/10.1093/nar/gkx407
Lyskov, S.; Gray, J. J. Nucleic Acids Res. 2008, 36, 233-238. DOI: https://doi.org/10.1093/nar/gkn216. DOI: https://doi.org/10.1093/nar/gkn216
McRee, D. E., in Elsevier eBooks. Practical Protein Crystallography, 1999. DOI: https://doi.org/10.1016/b978-0-12-486052-0.x5000-3. DOI: https://doi.org/10.1016/B978-012486052-0/50007-5
Morris, G. M.; Goodsell, D. S.; Pique, M. E.; Lindstrom, W.; Huey, R.; Forli, S.; Hart, W.; Halliday, S.; Belew, R.; Olson, A. User Guide. AutoDock Version 4.2. 2014. autodock.scripps.edu, https://autodock.scripps.edu/wp-content/uploads/sites/56/2021/10/AutoDock4.2.6_UserGuide.pdf, accessed in July 2023.
Protein Data Bank Protein Data Bank Contents Guide: Atomic Coordinate Entry Format Description. 2010. http://www.wwpdb.org/documentation/file-format-content/format33/v3.3.html, accessed in July 2023.
Huey, R.; Morris, G. M.; Olson, A. J.; Goodsell, D. S. J. Compl. Chem. 2007, 28, 1145-1152. DOI: https://doi.org/10.1002/jcc.20634. DOI: https://doi.org/10.1002/jcc.20634
Vieira, T. F.; Sousa, S. F. Appl. Sci. 2019, 9, 4538. DOI: https://doi.org/10.3390/app9214538. DOI: https://doi.org/10.3390/app9214538
Fischer, A.; Smieško, M.; Sellner, M.; Lill, M. A. J. Med. Chem. 2021, DOI: 2489-2500. DOI: https://doi.org/10.1021/acs.jmedchem.0c02227. DOI: https://doi.org/10.1021/acs.jmedchem.0c02227
García Nieto, J.M.; López Camacho, E.; García Godoy, M.J.; Nebro, A.J.; Durillo, J.J.; Aldana Montes, J.F., in ANTS 2016: 10th International Conference on Swarm Intelligence, Bruxelles, Belgique: Springer, 2016, 40-52. http://hdl.handle.net/10630/12124. DOI: https://doi.org/10.1007/978-3-319-44427-7_4
Golovko, T.; Min, R.; Lozovaya, N.; Falconer, C. J.; Yatsenko, N.; Tsintsadze, T.; Tsintsadze, V.; Ledent, C.; Harvey, R. J.; Belelli, D.; Lambert, J. J.; Rozov, A.; Burnashev, N. Cereb. Cortex. 2014. 25, 2440-2455. DOI: https://doi.org/10.1093/cercor/bhu045. DOI: https://doi.org/10.1093/cercor/bhu045
Forli, S.; Huey, R.; Pique, M. E.; Sanner, M. F.; Goodsell, D. S.; Olson, A. J. Nat. Prot. 2016. 11, 905-919. DOI: https://doi.org/10.1038/nprot.2016.051. DOI: https://doi.org/10.1038/nprot.2016.051
Coleman, R. G.; Carchia, M.; Sterling, T.; Irwin, J. J.; Shoichet, B. K. PloS One. 2013. 8, e75992. DOI: https://doi.org/10.1371/journal.pone.0075992. DOI: https://doi.org/10.1371/journal.pone.0075992
Friesner, R. A.; Banks, J. L.; Murphy, R. B.; Halgren, T. A.; Klicic, J. J.; Mainz, D. T.; Repasky, M. P.; Knoll, E. H.; Shelley, M.; Perry, J. K.; Shaw, D. E.; Francis, P.; Shenkin, P. S. J. Med. Chem. 2004, 47, 1739-1749. DOI: https://doi.org/10.1021/jm0306430. DOI: https://doi.org/10.1021/jm0306430
Thompson, M.A. Molecular Docking Using ArgusLab, an Efficient Shape-Based Search Algorithm and the A Score Scoring Function. ACS Meeting, Philadelphia. 2004.
Bitencourt-Ferreira, G.; de Azevedo, W. F., Methods Mol. Biol. 2019, 2053, 203-220. DOI: 10.1007/978-1-4939-9752-7_13. DOI: https://doi.org/10.1007/978-1-4939-9752-7_13
Thomsen, R.; Christensen, M. H. J. Med. Chem. 2006, 49, 3315-3321. DOI: https://doi.org/10.1021/jm051197e. DOI: https://doi.org/10.1021/jm051197e
Trott, O.; Olson, A. J. J. Comp. Chem. 2009, 31, 455-461. DOI: https://doi.org/10.1002/jcc.21334. DOI: https://doi.org/10.1002/jcc.21334
Jones, G.; Willett, P.; Glen, R. C.; Leach, A. R.; Taylor, R. J. Mol. Bio. 1997, 267, 727-748. DOI: https://doi.org/10.1006/jmbi.1996.0897. DOI: https://doi.org/10.1006/jmbi.1996.0897
Minibaeva, G.; Ivanova, A.; Polishchuk, P. J. Cheminform. 2023, 15, 102. DOI: https://doi.org/10.1186/s13321-023-00772-2. DOI: https://doi.org/10.1186/s13321-023-00772-2
Downloads
Additional Files
Published
Issue
Section
License
Copyright (c) 2024 A. Suárez-Alonso, A. Giacoman Martínez, E. F. Alarcón-Villaseñor, L. D. Herrera-Zúñiga, F. J. Alarcón-Aguilar

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.