Design and Synthesis of Barbiturates and Hydantoins with Multitarget Antidiabetic Effect☆
DOI:
https://doi.org/10.29356/jmcs.v68i4.2284Keywords:
Diabetes, Bioisosteres, Multitarget effect, Pharmacological consensus analysisAbstract
☆ Taking in part of the Master in Pharmacy thesis of S. Juárez-Cruz
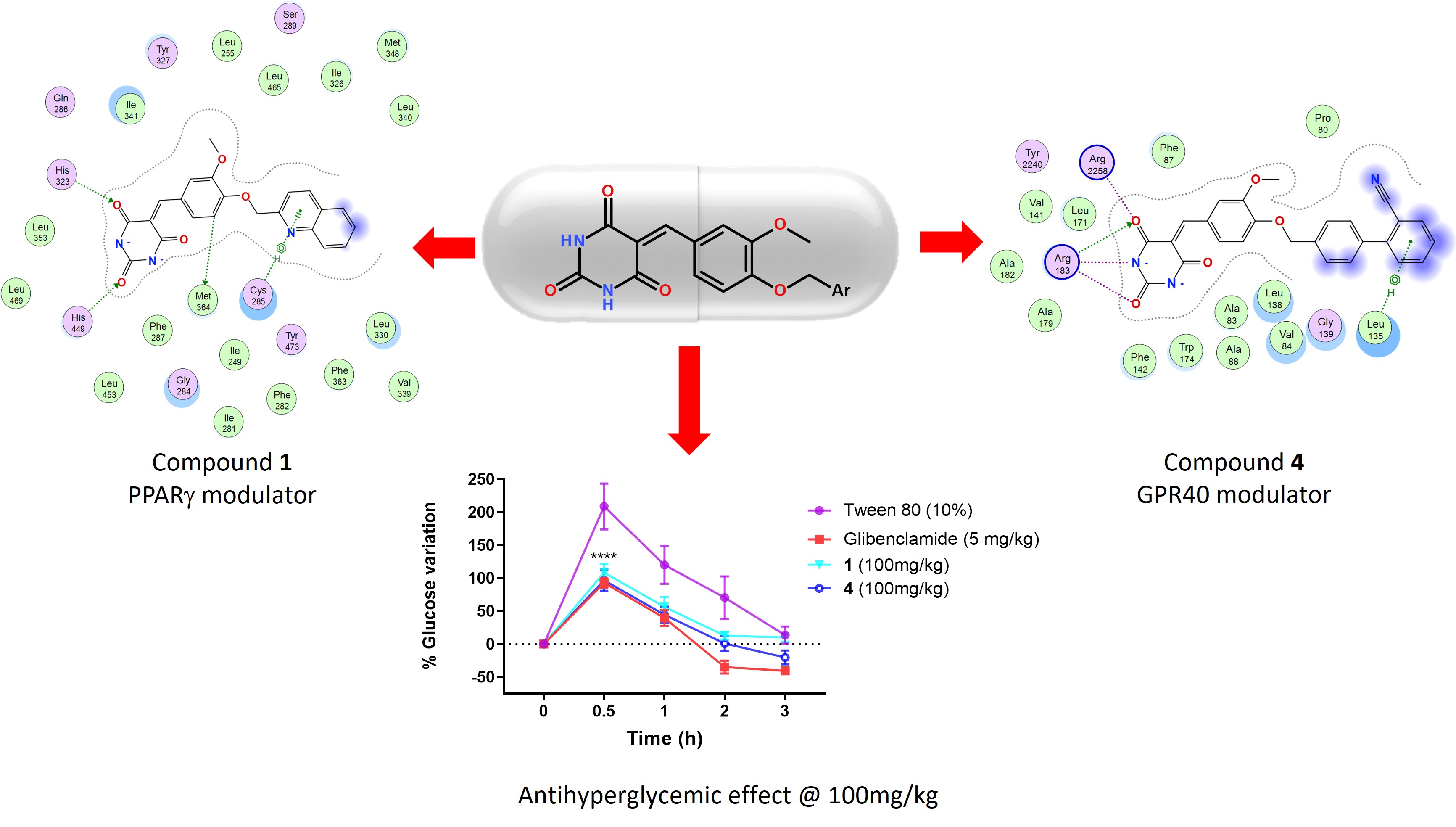
In current work, we prepared a series of ten 4-aryloxy-5-benzylidenebarbiturates and hydantoins as 1,3-thiazolidine-2,4-dione bioisosteres. An in silico pharmacological consensus analysis (PHACA) was conducted to assess the pharmacokinetic, pharmacodynamics, biopharmaceutical, and toxicological properties of compounds 1-10. The goal was to identify computationally safe hits using a color-coded system resembling a traffic light. The compounds identified as safe computational hits through PHACA were 1, 2, and 4 from the barbiturate series, which were then selected by in vitro assays targeting PPAR-γ, GPR40, and GLUT-4 gene expression. Additionally, these three compounds underwent in vivo evaluation through a glucose tolerance curve assay conducted on normoglycemic mice. Compounds 1 and 4 exhibited antihyperglycemic effects within the first thirty minutes post-administration. Molecular docking studies were conducted to clarify the dual effect and binding mode of compounds 1, 2 and 4 on PPAR-γ and GPR40. Compounds 1 and 4 exhibited robust in vitro and in vivo efficacy and could be considered as multitarget modulators with antidiabetic effect.
Resumen. En este trabajo se preparó una serie de diez 4-ariloxi-5-bencilidenobarbituratos e hidantoínas como bioisósteros de la 1,3-tiazolidina-2,4-diona. Se realizó un análisis de consenso farmacológico in silico (PHACA) para evaluar las propiedades farmacocinéticas, farmacodinámicas, biofarmacéuticas y toxicológicas de los compuestos 1-10. El objetivo era identificar hits computacionales seguros utilizando un sistema codificado por colores que se asemeja a un semáforo. Los compuestos identificados como hits computacionales seguros fueron 1, 2 y 4 de la serie de barbituratos, que se eligieron para ensayos in vitro dirigidos a la expresión génica de PPAR-γ, GPR40 y GLUT-4. Además, estos tres compuestos se sometieron a una evaluación in vivo mediante un ensayo de curva de tolerancia a la glucosa realizado en ratones normoglucémicos. Los compuestos 1 y 4 exhibieron efectos antihiperglucémicos dentro de los primeros treinta minutos posteriores a la administración. Se realizaron estudios de acoplamiento molecular para clarificar el efecto dual y el modo de unión de los compuestos 1, 2 y 4 en PPAR-γ y GPR40. Los compuestos 1 y 4 exhibieron una sólida eficacia in vitro e in vivo, por lo que pueden considerarse moduladores polifarmacológicos con efecto antidiabético.
Downloads
References
Mahboob, A.; Senevirathne, D.K.L.; Paul, P.; Nabi, F.; Khan, R.H.; Chaari, A. Int. J. Biol. Macromol. 2023, 225, 318-350. DOI: https://doi.org/10.1016/j.ijbiomac.2022.11.038.
Madrigal-Angulo, J.L.; Ménez-Guerrero, C.; Estrada-Soto, S.; Ramírez-Espinosa, J.J.; Almanza-Pérez, J.C.; León-Rivera, I.; Hernández-Núñez, E.; Aguirre-Vidal, Y.; Flores-León, C.D.; Aguayo-Ortíz, R.; Navarrete-Vazquez G. Bioorg. Med. Chem. Lett. 2022, 70, 128804. DOI: https://doi.org/10.1016/j.bmcl.2022.128804.
Hidalgo-Figueroa, S.; Rodríguez-Luévano, A.; Almanza-Pérez, J.C.; Giacoman-Martínez, A.; Ortiz-Andrade, R.; León-Rivera, I.; Navarrete-Vázquez, G. Eur. J. Pharmacol. 2021, 907,174244. DOI: https://doi.org/10.1016/j.ejphar.2021.174244.
Ren, Q.; Fan, Y.; Yang, L.; Shan, M.; Shi, W.; Qian, H. Expert. Opin. Ther. Pat. 2023, 33, 565-577. DOI: https://doi.org/10.1080/13543776.2023.2272649.
Saldívar-González, F.I.; Navarrete-Vázquez, G.; Medina-Franco, J.L. Front. Pharmacol. 2023, 14:1276444. DOI: https://doi.org/10.3389/fphar.2023.1276444.
Colín-Lozano, B.; Estrada-Soto, S.; Chávez-Silva, F.; Gutiérrez-Hernández, A.; Cerón-Romero, L.; Giacoman-Martínez, A.; Almanza-Pérez, J.C.; Hernández-Núñez, E.; Wang, Z.; Xie, X.; Cappiello, M.; Balestri, F.; Mura, U.; Navarrete-Vazquez, G. Molecules. 2018, 23, 340. DOI: https://doi.org/10.3390/molecules23020340.
Giacoman-Martínez, A.; Alarcón-Aguilar, F.J.; Zamilpa, A.; Hidalgo-Figueroa, S.; Navarrete-Vázquez, G.; García-Macedo, R.; Román-Ramos, R.; Almanza-Pérez, J.C. Planta Med. 2019, 85, 412-423. DOI: https://doi.org/10.1055/a-0824-1316.
Giacoman-Martínez, A.; Alarcón-Aguilar, F.J.; Zamilpa, A.; Huang, F.; Romero-Nava, R.; Román-Ramos, R.; Almanza-Pérez, J.C. Can J. Physiol. Pharmacol. 2021, 99, 935-942. DOI: https://doi.org/10.1139/cjpp-2021-0027.
Rosiles-Alanis, W.; Zamilpa, A.; García-Macedo, R.; Zavala-Sánchez, M.A.; Hidalgo-Figueroa, S.; Mora-Ramiro, B.; Román-Ramos, R.; Estrada-Soto, S.E.; Almanza-Perez, J.C. J. Med. Food. 2022, 25, 588-596. DOI: https://doi.org/10.1089/jmf.2021.0071.
Estrada-Soto, S.; Ornelas-Mendoza, K.; Navarrete-Vázquez, G.; Chávez-Silva, F.; Almanza-Pérez, J.C.; Villalobos-Molina, R., Ortiz-Barragán, E.; Loza-Rodríguez, H.; Rivera-Leyva, J.C.; Flores-Flores, A.; Perea-Arango, I.; Rodríguez-Carpena, J.G.; Ávila-Villarreal, G. Pharmaceuticals. 2023, 16, 535. DOI: https://doi.org/10.3390/ph16040535.
Molecular Operating Environment (MOE). Chemical Computing group ULC; 910-1010 Sherbooke St. West, Montreal, QC, Canada, 2024; Available online: http://www.chemcomp.com, accessed in January 2024.
Srivastava, A.; Yano, J.; Hirozane, Y.; Kefala, G.; Gruswitz, F.; Snell, G.; Lane, W.; Ivetac, A.; Aertgeerts, K.; Nguyen, J.; Jennings, A.; Okada, K. Nature. 2014, 513, 124-127. DOI: https://doi.org/10.1038/nature13494.
Liberato, M.V.; Nascimento, A.S.; Ayers, S.D.; Lin, J.Z.; Cvoro, A.; Silveira, R.L.; Martínez, L.; Souza, P.C.; Saidemberg, D.; Deng, T.; Amato, A.A.; Togashi, M.; Hsueh, W.A.; Phillips, K.; Palma, M.S.; Neves, F.A.; Skaf, M.S.; Webb, P.; Polikarpov, I. PLoS One. 2012, 7, e36297. DOI: https://doi.org/10.1371/journal.pone.0036297.
Schrödinger, L., & DeLano, W. PyMOL. Retrieved from http://www.pymol.org/pymol, accessed in 2024.
Domínguez-Mendoza, E.A.; Galván-Ciprés, Y.; Martínez-Miranda, J.; Miranda-González, C.; Colín-Lozano, B.; Hernández-Núñez, E.; Hernández-Bolio, G.I.; Palomino-Hernández, O.; Navarrete-Vazquez, G. Molecules. 2021, 26, 799. DOI: https://doi.org/10.3390/molecules26040799.
Molinspiration Cheminformatics free web services, Available online: https://www.molinspiration.com, accessed in February 2023.
Xiong, G., Wu, Z., Yi, J., Fu, L., Yang, Z., Hsieh, C., Yin, M., Zeng, X., Wu, C., Lu, A., Chen, X., Hou, T., Cao, D. Nucleic Acids Res. 2021, 49, W5-W14. ADMETLab 2.0 Available online: https://admetmesh.scbdd.com, accessed March 2022. DOI: https://doi.org/10.1093/nar/gkab255
Rudik, A.V.; Dmitriev, A.V.; Lagunin, A.A.; Filimonov, D.A.; Poroikov, V.V. ACS Omega. 2023, 8, 45774-45778. DOI: 10.1021/acsomega.3c06119. MetaTox 2.0 Available online: https://www.way2drug.com/metatox , accessed in December 2023. DOI: https://doi.org/10.1021/acsomega.3c06119
Diaza, R.G.; Manganelli, S.; Esposito, A.; Roncaglioni, A.; Manganaro, A.; Benfenati, E. SAR QSAR Environ Res. 2015, 26, 1-27. DOI: https://doi.org/10.1080/1062936X.2014.977819.
Bonatto, V.; Lameiro, R.F.; Rocho, F.R.; Lameira, J.; Leitão, A.; Montanari, C.A. RSC Med. Chem. 2022, 14, 201-217. DOI: https://doi.org/10.1039/d2md00204c.
Korbecki, J.; Bobiński, R.; Dutka, M. Inflamm. Res. 2019, 68, 443-458. DOI: https://doi.org/10.1007/s00011-019-01231-1.
Lückmann, M.; Trauelsen, M.; Bentsen, M.A.; Nissen, T.A.D.; Martins, J.; Fallah, Z.; Nygaard, M.M.; Papaleo, E.; Lindorff-Larsen, K.; Schwartz, T.W.; Frimurer, T.M. Proc. Natl. Acad. Sci. U S A. 2019, 116, 7123-7128. DOI: https://doi.org/10.1073/pnas.1811066116.

Downloads
Additional Files
Published
Issue
Section
License
Copyright (c) 2024 Gabriel Navarrete-Vazquez, Samantha Juárez-Cruz, Samuel Estrada-Soto, Blanca Colin-Lozano, Hugo Marquina-Rodriguez, Thalía Delgado-Aguilar, Carlos Martínez-Conde, Abraham Gutiérrez-Hernández, Emanuel Hernández-Núñez, Abraham Giacoman-Martínez, Julio Cesar Almanza-Pérez

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.