Biopanning Phage Display Libraries in Homogeneous Solution for Identification of Biomineralization Peptides of TiO₂
DOI:
https://doi.org/10.29356/jmcs.v68i4.2262Keywords:
Phage display, peptides, titanium oxide, biopanning, biomineralizationAbstract
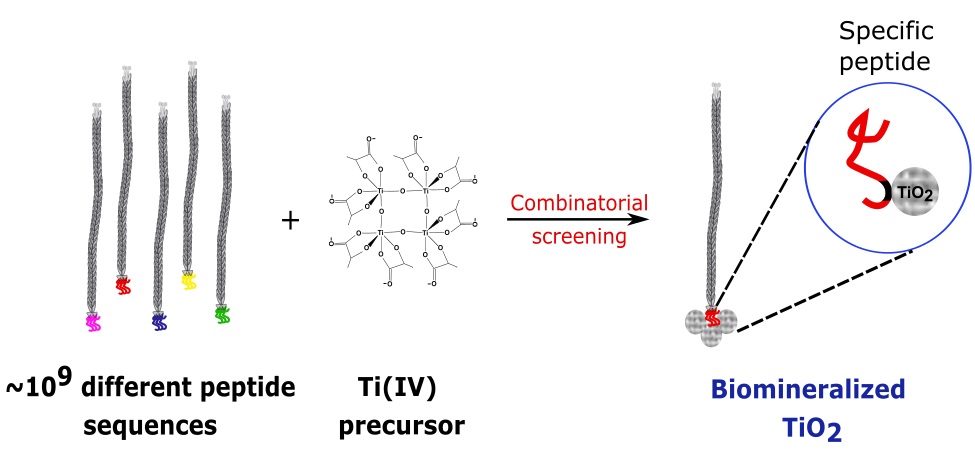
Peptides and proteins rich in positively charged residues have been the most frequently reported for TiO2 biomineralization since their identification is based on peptide screening on its negatively charged surface. To achieve optimum interaction of the peptides with the biomimetic synthesis precursors rather than interaction with the final product, in this work, a selection of peptides with biomineralization activity was proposed by performing a biopanning directly on the precursor Titanium(IV) bis(ammonium lactate) dihydroxide (TiBALDH). Using two phage display libraries (12- and 7-mer) in different buffer systems, four possible sequences with biomineralization activity of TiO2 were identified: TNWQALAYMQRH (TN), ENHWSLSTLMSS (EN), GLHTSATNLYLH (GL), TWYPNRPPILEL (TW). The selection of buffer and concentration of TiBALDH were vital for a reliable identification. Synthetic peptides with sequences TN and EN, were selected for in vitro biomineralization of TiO2. Both peptides were able to form anatase nanoparticles at room temperature. However, the EN sequence showed lower activity than TN, specially in acetate buffer, requiring a higher concentration to initiate biomineralization. These changes in reactivity can be attributed principally to different states of protonation of the residues mainly due to the glutamic acid in EN. Although the secondary structure determined by circular dichroism results in disordered chains, a common motif could be identified between the two peptides -pol-pol-W-pol-x-x-x-x-M-, where the W and M residues match. The results provide new possibilities for using combinatorial techniques to find new biological templates for nanomaterial synthesis.
Resumen. Péptidos y proteínas ricas en residuos con carga positiva han sido frecuentemente reportados para la biomineralización de TiO2, ya que su identificación se basa en la detección de péptidos sobre su superficie con carga negativa. Para alcanzar una interacción óptima del péptido con el precursor biomimético, en lugar de la interacción con el producto final, en este trabajo, se propuso realizar un biotamizado empleando el precursor dihidroxilactatotitanato(IV) de bis-amonio (TiBALDH) para seleccionar péptidos con actividad de biomineralización. Empleamos dos librerías de fago desplegado (12 y 7 residuos) en diferentes soluciones amortiguadoras, identificando cuatro posibles secuencias con actividad biomineralizante de TiO2: TNWQALAYMQRH (TN), ENHWSLSTLMSS (EN), GLHTSATNLYLH (GL), TWYPNRPPILEL (TW). La elección del amortiguador y la concentración de TiBALDH fueron vitales para una selección confiable. Los péptidos sintéticos TN y EN, fueron escogidos para la biomineralización de TiO2 in vitro. Ambos péptidos fueron capaces de formar nanopartículas de anatasa a temperatura ambiente, sin embargo, la secuencia EN mostró menor actividad que TN, especialmente en amortiguador de acetatos, requiriendo una concentración mayor para iniciar la biomineralización. Estas diferencias de reactividad pueden ser atribuidas principlamente a los estados de protonación de los residuos de ácido glutámico en EN. Aunque la estructura secundaria determinada por dicroísmo circular mostró cadenas desordenadas, se identificó un motivo común entre los dos péptidos—pol-pol-W-pol-x-x-x-x-M-, donde los aminoácidos W y M coinciden. Los resultados abren nuevas posibilidades para usar técnicas combinatorias para hallar nuevas plantillas biológicas para la síntesis de nanomateriales.
Downloads
References
Heintze, C.; Babenko, I.; Suchanova, J. Z.; Skeffington, A.; Friedrich, B. M.; Kröger, N. Proc. Natl. Acad. Sci. 2022, 119, e2211549119. DOI: https://doi.org/10.1073/pnas.2211549119.
Otter, L.M.; Eder, K.; Kilburn, M.R. et al. Nat. Commun. 2023, 14, 2254. DOI: https://doi.org/10.1038/s41467-023-37814-0.
Sharma, V.; Srinivasan, A.; Nikolajeff, F.; Kumar, S. Acta Biomaterialia. 2021, 120, 20-37. DOI: https://doi.org/10.1016/j.actbio.2020.04.049.
Liu, F.; Shah, D. S.; Gadd, G. M. Curr. Biol. 2021, 31, 358-368. DOI: https://doi.org/10.1016/j.cub.2020.10.044.
Buettner, K. M.; Valentine, A. M. Chem. Rev. 2011, 112, 1863-1881. DOI: https://doi.org/10.1021/cr1002886
Jha, A. K.; Prasad, K.; Kulkarni, A. R. Colloids Surf. B. 2009, 71, 226-229. DOI: https://doi.org/10.1016/j.colsurfb.2009.02.007.
Lang, Y.; Monte, F. D.; Rodriguez, B. J.; Dockery, P.; Finn, D. P.; Pandit, A. Sci. Rep. 2013, 3, 3205. DOI: 10.1038/srep03205. DOI: https://doi.org/10.1038/srep03205
Smith G. P.; Petrenko V. A. Chem. Rev. 1997, 97, 391–410. DOI: https://doi.org/10.1021/cr960065d.
Bratkovič T. Cell. Mol. Life Sci. 2010, 67, 749-767. DOI: https://doi.org/10.1007/s00018-009-0192-2.
10. Rosant, C.; Avalle, B.; Larcher, D.; Dupont, L.; Fribouleta, A.; Tarascon, J.-M. Energy Environ. Sci. 2012, 5, 9936–9943. DOI: https://doi.org/10.1039/C2EE22234E.
Dickerson, M. B.; Jones, S. E.; Cai, Y.; Ahmad, G.; Naik, R. R.; Kröger, N.; Sandhage, K. H. Chem. Mater. 2008, 20, 1578-1584. DOI: https://doi.org/10.1021/cm071515t.
Sun, Y.; Tan, J.; Wu, B.; Wang, J.; Qu, S.; Weng, J.; Feng, B. Colloids Surf. B. 2016, 146, 307-317. DOI: https://doi.org/10.1016/j.colsurfb.2016.06.032.
Chen, H.; Su, X.; Neoh, K.-G.; Choe, W.-S. Anal. Chem. 2006, 78, 4872-4879. DOI: https://doi.org/10.1021/ac0603025.
Park, S.; Lee, H.; Lee, S.-Y. Dalton Trans. 2013, 42, 13817-13820. DOI: https://doi.org/10.1039/C3DT51040A.
Liu, C.; Jiang, Z.; Tong, Z.; Li, Y.; Yang, D. RSC Adv. 2014, 4, 434-441. DOI: https://doi.org/10.1039/C3RA44630A.
Puddu, V.; Slocik, J. M.; Naik, R. R.; Perry, C. C. Langmuir. 2013, 29, 9464–9472. DOI: https://doi.org/10.1021/la401777x.
Hellner, B.; Stegmann, A. E.; Pushpavanam, K.; Bailey, M. J.; Baneyx F. Langmuir. 2020, 36, 8503–8510. DOI: https://doi.org/10.1021/acs.langmuir.0c01108.
Choi, N.; Tan, L.; Jang, J.-r.; Um, Y.M.; Yoo, P.J.; Choe, W.-S. J. Inorg. Biochem. 2012, 115, 20-27. DOI: https://doi.org/10.1016/j.jinorgbio.2012.05.011.
Zelechowska, K.; Karczewska-Golec, J.; Karczewski, J.; Los, M.; Klonkowski, A. M.; Wegrzyn, G.; Golec, P. Bioconjugate Chem. 2016, 27, 1999–2006. DOI: https://doi.org/10.1021/acs.bioconjchem.6b00196.
Maeda, Y.; Javid, N.; Duncan, K.; Birchall, L.; Gibson, K. F.; Cannon, D.; Kanetsuki, Y.; Knapp, C.; Tuttle, T.; Ulijn, R. V.; Matsui. H. J. Am. Chem. Soc. 2014, 136, 15893−15896. DOI: https://doi.org/10.1021/ja509393p.
Seisenbaeva, G. A.; Daniel, G.; Nedelec, J.-M.; Kessler, V. G. Nanoscale. 2013, 5, 3330. DOI: https://doi.org/10.1039/C3NR34068F.
Hernández-Gordillo, A.; Hernández-Arana, A.; Campero-Celis, A.; Vera-Robles, L.I. RSC Adv. 2019, 9, 34559-34566. DOI: https://doi.org/10.1039/C9RA05923G.
Moharir, A.V.; Sarma, V.A.K.; Krishna Murti G.S.R. Microchem. J. 1972, 17, 167-172. DOI: https://doi.org/10.1016/0026-265X(72)90169-5.
Hu, Y.-F.; Gao, X.-C.; Xu, T.-Q.; Dun, Z.; Yu, X.-L. Comb. Chem. High Throughput Screening. 2016, 19, 283-289. DOI: https://doi.org/10.2174/1386207319666160316122106
Vodnik, M.; Zager, U.; Strukelj, B.; Lunder, M. Molecules. 2011, 16, 790-817. DOI: https://doi.org/10.3390/molecules16010790.
Majerova, P.; Hanes, J.; Olesova, D.; Sinsky, J.; Pilipcinec, E.; Kovac, A. Molecules. 2020, 25, 874. DOI: https://doi.org/10.3390/molecules25040874.
Nemudraya, A. A.; Kuligina, E.V .; Ilyichev, A. A.; Fomin, A. S.; Stepanov, G. A.; Savelyeva, A.V.; Koval, O.A.; Richter, V.A. Oncol. Lett. 2016, 12, 4547-4555. DOI: https://doi.org/10.3892/ol.2016.5266.
Zhang, H.; Guo, Z.; He, B.; Dai, W.; Zhang, H.; Wang, X.; Zhang, Q. Adv. Healthc. Mater. 2018, 7, 1800269. DOI: https://doi.org/10.1002/adhm.201800269.
Clustal Omega https://www.uniprot.org/align/clustalo-R20220708-213026-0610-21074307-p1m, accessed in July 2022.
http://protcalc.sourceforge.net/, accessed in January 2022
Rucker, A.L.; Creamer T.P. Protein Sci. 2002, 11, 980-985 doi: 10.1110/ps.4550102. DOI: https://doi.org/10.1110/ps.4550102
Reed, J,; Reed T. A. Anal Biochem. 1997, 254, 36-40 https://doi.org/10.1006/abio.1997.2355.
(a)Whitmore, L.; Wallace, B. A. Nucleic Acids Res. 2004, 32, W668–W673. DOI: https://doi.org/10.1093/nar/gkh371. (b)Whitmore, L.; Wallace, B.A. Biopolymers 2008, 89, 392–400. DOI: https://doi.org/10.1002/bip.20853. (c)http://www.cryst.bbk.ac.uk/cdweb, accessed in June 2022. (d)Sreerama, N.; Woody, R.W. Anal. Biochem. 2000, 287, 252–260.
Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.-H.; Goto, Y.; M.; Réfrégiers, Kardos, J. Nucleic Acids Research 2018, 46, W315 W322. DOI: https://doi.org/10.1093/nar/gky497. http://bestsel.elte.hu.
Hernández-Gordillo, A.; Hernández-Arana, A.; Campero, A.; Vera-Robles, L.I. Langmuir. 2014, 30, 4084-4093. DOI: https://doi.org/10.1021/la500203k.

Downloads
Published
Issue
Section
License
Copyright (c) 2024 Irais Vera-Robles, Armin Hernández-Arana, Andrés Hernández-Arana

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.