2-Oxoquinoline Arylaminothiazole Derivatives in Identifying Novel Potential Anticancer Agents by Applying 3D-QSAR, Docking, and Molecular Dynamics Simulation Studies
DOI:
https://doi.org/10.29356/jmcs.v66i1.1578Keywords:
3D-QSAR, molecular docking, MD simulation, quinoline, cancerAbstract
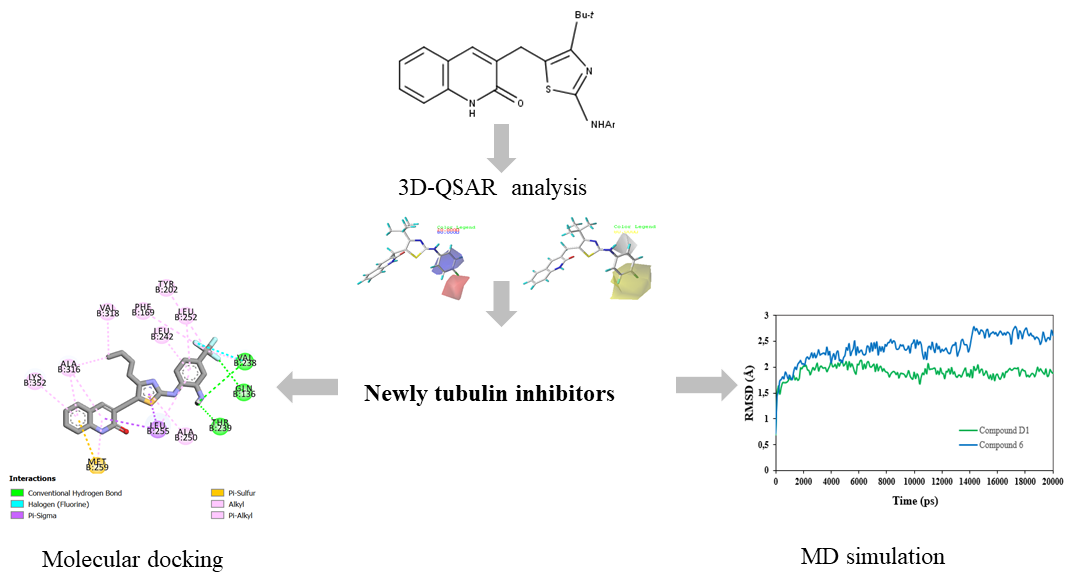
Abstract. Tubulin plays an indispensable role in regulating various important cellular processes. Recently, it is known as a hopeful therapeutic target for the rapid division of cancer cells. Novel series of 2-oxoquinoline arylaminothiazole derivatives have been recently identified as promising tubulin inhibitors with potent cytotoxicity activity against HeLa cancer cell line. In this study, a 3D-QSAR approach by using CoMFA and CoMSIA techniques was applied to the reported derivatives to understand their pharmacological essentiality contributing to the tubulin inhibition activity and selectivity. The optimum CoMFA and CoMSIA models were found to have significant statistical reliability and high predictive ability after internal and external validation. By analyzing the contour maps, the electrostatic and hydrophobic interactions were found to be crucial for improving the inhibitory activity and four novel tubulin inhibitors (Compounds D1, D2, D3, and D4) were designed based on the validated 3D-QSAR models. Moreover, the docking findings showed that residues Gln136, Val238, Thr239, Asn167, Val 318 and Ala 316 played important roles for quinoline binding to tubulin. Among the newly designed compounds, compound D1 with the highest total scoring was subjected to detailed molecular dynamics (MD) simulation and compared to the most active compound. The conformational stability of compound D1 complexed with tubulin protein was confirmed by a 50-ns molecular dynamics simulation, which was congruent with molecular docking.
Resumen. La tubulina juega un papel indispensable en la regulación de varios procesos celulares importantes. Recientemente, se le ha reconicodo como un agente promisorio para atacar la rápida división de las células cancerosas. Últimamente se ha identificado una nueva serie de derivados de arilaminotiazo-2-oxoquinolina como potenciales inhibidores de la tubulina, con una elevada actividad citotóxica contra la línea celular de cáncer HeLa. En este estudio, se aplicó a los derivados informados un estudio 3D-QSAR mediante el uso de técnicas CoMFA y CoMSIA para comprender los factores farmacológicos que contribuyen a la actividad como inhibidor y selectivo de la tubulina. Se encontró que los modelos CoMFA y CoMSIA óptimos tienen una confiabilidad estadística significativa y una alta capacidad predictiva después de la validación interna y externa. Al analizar los mapas de contorno, se descubrió que las interacciones electrostáticas e hidrófobas eran cruciales para mejorar la actividad inhibidora y se diseñaron cuatro nuevos inhibidores de la tubulina (compuestos D1, D2, D3 y D4) basados en los modelos 3D-QSAR validados. Además, los hallazgos de acoplamiento mostraron que los residuos Gln136, Val238, Thr239, Asn167, Val 318 y Ala 316 desempeñaron papeles importantes en la unión de la quinolina a la tubulina. Entre los compuestos de nuevo diseño, el compuesto D1 con la puntuación total más alta se sometió a una simulación detallada de dinámica molecular (MD) y se comparó con el compuesto más activo. La estabilidad conformacional del compuesto D1 unido a la proteína tubulina se confirmó mediante una simulación de dinámica molecular de 50 ns, que fue congruente con el acoplamiento molecular.
Downloads
References
Siegel, R. L.; Miller, K. D.; Jemal, A. CA. Cancer J. Clin. 2019, 69, 7–34. DOI: https://doi.org/10.3322/caac.21551
Sana, S.; Tokala, R.; Bajaj, D. M.; Nagesh, N.; Bokara, K. K.; Kiranmai, G; Lakshmi, U. J.; Vadlamani, S.; Talla, V. Shankaraiah, N. Bioorg. Chem. 2019, 93, 103317. DOI: https://doi.org/10.1016/j.bioorg.2019.103317
Kaur, R.; Kaur, G.; Gill, R. K.; Soni, R.; Bariwal, J. Eur. J. Med. Chem. 2014, 87, 89-124. DOI: https://doi.org/10.1016/j.ejmech.2014.09.051
Liu, Y.-N.; Wang, J.-J.; Ji, Y.-T.; Zhao, G.-D.; Tang, L.-Q.; Zhang, C.-M.; Guo, X.-L.; Liu, Z.-P. J. Med. Chem. 2016, 59, 5341–5355. DOI: https://doi.org/10.1021/acs.jmedchem.6b00071
Stanton, R. A.; Gernert, K. M.; Nettles, J. H.; Aneja, R. Med. Res. Rev. 2011, 31, 443–481. https://doi.org/10.1002/med.20242
Mukhtar, E.; Adhami, V. M.; Mukhtar, H. Mol. Cancer Ther. 2014, 13, 275–284. DOI: https://doi.org/10.1158/1535-7163.MCT-13-0791
Bates, D.; Eastman, A. Br. J. Clin. Pharmacol. 2017, 83, 255–268. DOI: https://doi.org/10.1111/bcp.13126
Batran, R. Z.; Kassem, A. F.; Abbas, E. M. H.; Elseginy, S. A.; Mounier, M. M. Med. Chem. 2018, 26, 3474–3490. DOI: https://doi.org/10.1016/j.bmc.2018.05.022
Vuuren, R. J.; Visagie, M. H.; Theron, A. E.; Joubert, A. M. Cancer Chemother Pharmacol. 2015, 76, 1101–1112. https://doi.org/10.1007/s00280-015-2903-8
Jordan, M. A. Curr. Med. Chem. 2002, 2, 1-17. DOI: https://doi.org/10.2174/1568011023354290
Ibrahim, T. S.; Hawwas, M. M.; Malebari, A. M.; Taher, E. S.; Omar, A. M.; O’Boyle, N. M.; McLoughlin, E.; Abdel-Samii, Z. K. P. Pharmaceuticals. 2020, 13, 393. DOI: https://doi.org/10.3390/ph13110393
El-Naggar, A. M.; Eissa, I. H.; Belal, A.; EL-Sayed, A. A. RSC Adv. 2020, 10, 2791–2811. DOI: https://doi.org/10.1039/c9ra10094f
Thiyagamurthy, P.; Teja, C.; Naresh, K.; Gomathi, K.; Khan, F-R. N. Med. Chem. Res. 2020, 29, 1882-1901. DOI: https://doi.org/10.1007/s00044-020-02606-4
Kaur, R.; Kumar, K. S. Eur. J. Med. Chem. 2021, 215, 113220. DOI: https://doi.org/10.1016/j.ejmech.2021.113220
Mahajan, P.; Nikam, M.; Asrondkar, A.; Bobade, A.; Gill, C. J. Heterocyclic Chem. 2016, 000, 8. DOI: https://doi.org/10.1002/jhet.2722
Uddin, A.; Chawla, M.; Irfan, I.; Mahajan, S.; Singh, S.; Abid, M. RSC Med. Chem. 2021, 12, 24-42. DOI: https://doi.org/10.1039/d0md00244e
Katariya, K. D.; Shah, S. R.; Reddy, D. Bioorg. Chem. 2020, 94, 103406. DOI: https://doi.org/10.1016/j.bioorg.2019.103406
Fang, Y.; Wu, Z.; Xiao, M.; Li Wei, L.; Li, K.; Tang, Y.; Ye, J.; Xiang, J.; Hu, A. Bioorg. Chem. 2021, 106, 104469. DOI: https://doi.org/10.1016/j.bioorg.2020.104469
S TRIPOS Associates, Inc. Sybyl-X Molecular Modeling Software Packages., Version X-2.0. 2012
Clark, M.; Cramer, R. D.; Van Opdenbosch, N. J. Comput. Chem. 1989, 10, 982–1012. DOI: https://doi.org/10.1002/jcc.540100804
Purcell, W. P.; Singer, J. A. J. Chem. Eng. Data. 1967, 12, 235–246. DOI: https://doi.org/10.1021/je60033a020
El Khatabi, K.; Aanouz, I.; El-mernissi, R.; Khaldan, A.; Ajana, M. A.; Bouachrine, M.; Lakhlifi, T. Orbital Electron. J. Chem. 2020, 12, 172–181. DOI: https://doi.org/10.17807/orbital.v12i4.1467
Cramer, R. D.; Patterson, D. E.; Bunce, J. D. J. Am. Chem. Soc. 1988, 110, 5959–5967. DOI: https://doi.org/10.1021/ja00226a005
Klebe, G.; Abraham, U.; Mietzner, T. J. Med. Chem. 1994, 37, 4130–4146. DOI: https://doi.org/10.1021/jm00050a010
Ståhle, L.; Wold, S. Prog. Med. Chem. 1988, 25, 291–338. DOI: https://doi.org/10.1016/S0079-6468(08)70281-9
EL-Mernissi, R.; El Khatabi, K.; Khaldan, A.; Ajana, M. A.; Bouachrine, M.; Lakhlifi, T. J. Mater. Environ. Sci. 2020, 11, 952-962.
Golbraikh, A.; Tropsha, A. J. Mol. Graph. Model. 2002, 20, 269–276. DOI: https://doi.org/10.1016/S1093-3263(01)00123-1
Tropsha, A.; Gramatica, P.; Gombar, V. QSAR Comb. Sci. 2003, 22, 69–77. DOI: https://doi.org/10.1002/qsar.200390007
Baroni, M.; Clementi, S.; Cruciani, G.; Costantino, G.; Riganelli, D.; Oberrauch, E. J. Chemom. 1992, 6, 347–356. DOI: https://doi.org/10.1002/cem.1180060605
Rücker, C.; Rücker, G.; Meringer, M. J. Chem. Inf. Model. 2007, 47, 2345–2357. DOI: https://doi.org/10.1021/ci700157b
Discovery Studio Predictive Science Application | Dassault Systèmes BIOVIA https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/ (accessed Feb 6, 2020).
DeLano, W. L. DeLano. PyMOL Mol. Graph. Syst. DeLano Sci. San Carlos CA USA 2002.
Kaminski, G. A.; Friesner, R. A.; Tirado-Rives, J.; Jorgensen, W. L. J. Phys. Chem. B. 2001, 105, 6474–6487. DOI: https://doi.org/10.1021/jp003919d
Schrödinger Release 2020–1: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2020. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2020.
Nam, K.; Gao, J.; York, D. M. J. Chem. Theory Comput. 2005, 1, 2–13. DOI: https://doi.org/10.1021/ct049941i

Downloads
Published
Issue
Section
License
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.